Brain decoding with SVM#
Support vector machines#
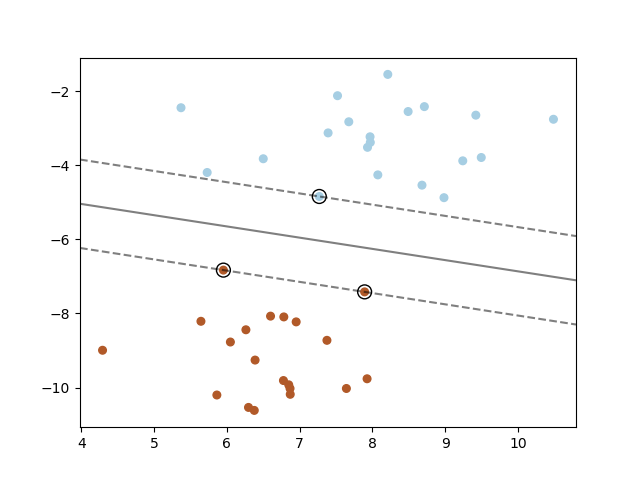
Fig. 4 A SVM aims at finding an optimal hyperplane to separate two classes in high-dimensional space, while maximizing the margin. Image from the scikit-learn SVM documentation under BSD 3-Clause license.#
We are going to train a support vector machine (SVM) classifier for brain decoding on the Haxby dataset. SVM is often successful in high dimensional spaces, and it is a popular technique in neuroimaging.
In the SVM algorithm, we plot each data item as a point in N-dimensional space where N depends on the number of features that distinctly classify the data points (e.g. when the number of features is 3 the hyperplane becomes a two-dimensional plane.). The objective here is finding a hyperplane (decision boundaries that help classify the data points) with the maximum margin (i.e the maximum distance between data points of both classes). Data points falling on either side of the hyperplane can be attributed to different classes.
The scikit-learn documentation contains a detailed description of different variants of SVM, as well as example of applications with simple datasets.
Getting the data#
We are going to download the dataset from Haxby and colleagues (2001) [HGF+01]. You can check section An overview of the Haxby dataset for more details on that dataset. Here we are going to quickly download it, and prepare it for machine learning applications with a set of predictive variable, the brain time series X, and a dependent variable, the annotation on cognition y.
import os
import warnings
warnings.filterwarnings(action='ignore')
from nilearn import datasets
# We are fetching the data for subject 4
data_dir = os.path.join('..', 'data')
sub_no = 4
haxby_dataset = datasets.fetch_haxby(subjects=[sub_no], fetch_stimuli=True, data_dir=data_dir)
func_file = haxby_dataset.func[0]
# mask the data
from nilearn.maskers import NiftiMasker
mask_filename = haxby_dataset.mask_vt[0]
masker = NiftiMasker(mask_img=mask_filename, standardize=True, detrend=True)
X = masker.fit_transform(func_file)
# cognitive annotations
import pandas as pd
behavioral = pd.read_csv(haxby_dataset.session_target[0], delimiter=' ')
y = behavioral['labels']
Let’s check the size of X and y:
categories = y.unique()
print(categories)
print(y.shape)
print(X.shape)
['rest' 'face' 'chair' 'scissors' 'shoe' 'scrambledpix' 'house' 'cat'
'bottle']
(1452,)
(1452, 675)
So we have 1452 time points, with one cognitive annotations each, and for each time point we have recordings of fMRI activity across 675 voxels. We can also see that the cognitive annotations span 9 different categories.
Training a model#
We are going to start by splitting our dataset between train and test. We will keep 20% of the time points as test, and then set up a 10 fold cross validation for training/validation.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
Now we can initialize a SVM classifier, and train it:
from sklearn.svm import SVC
model_svm = SVC(random_state=0, kernel='linear', C=1)
model_svm.fit(X_train, y_train)
SVC(C=1, kernel='linear', random_state=0)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
SVC(C=1, kernel='linear', random_state=0)
Assessing performance#
Let’s check the accuracy of the prediction on the training set:
from sklearn.metrics import classification_report
y_train_pred = model_svm.predict(X_train)
print(classification_report(y_train, y_train_pred))
precision recall f1-score support
bottle 1.00 1.00 1.00 85
cat 1.00 1.00 1.00 88
chair 1.00 1.00 1.00 90
face 1.00 1.00 1.00 81
house 1.00 1.00 1.00 91
rest 1.00 1.00 1.00 471
scissors 1.00 1.00 1.00 81
scrambledpix 1.00 1.00 1.00 90
shoe 1.00 1.00 1.00 84
accuracy 1.00 1161
macro avg 1.00 1.00 1.00 1161
weighted avg 1.00 1.00 1.00 1161
This is dangerously high. Let’s check on the test set:
y_test_pred = model_svm.predict(X_test)
print(classification_report(y_test, y_test_pred))
precision recall f1-score support
bottle 0.72 0.78 0.75 23
cat 0.70 0.70 0.70 20
chair 0.74 0.78 0.76 18
face 0.89 0.93 0.91 27
house 0.93 0.82 0.88 17
rest 0.91 0.89 0.90 117
scissors 0.84 0.78 0.81 27
scrambledpix 0.85 0.94 0.89 18
shoe 0.72 0.75 0.73 24
accuracy 0.84 291
macro avg 0.81 0.82 0.81 291
weighted avg 0.84 0.84 0.84 291
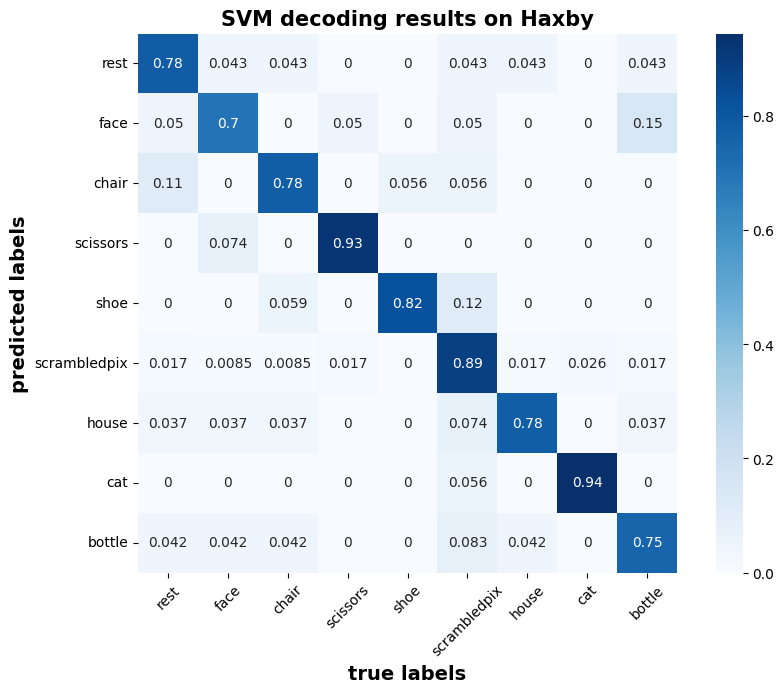
We can have a look at the confusion matrix:
# confusion matrix
import sys
import numpy as np
from sklearn.metrics import confusion_matrix
sys.path.append('../src')
import visualization
cm_svm = confusion_matrix(y_test, y_test_pred)
model_conf_matrix = cm_svm.astype('float') / cm_svm.sum(axis=1)[:, np.newaxis]
visualization.conf_matrix(model_conf_matrix,
categories,
title='SVM decoding results on Haxby')
Visualizing the weights#
Finally we can visualize the weights of the (linear) classifier to see which brain region seem to impact most the decision, for example for faces:
from nilearn import plotting
# first row of coef_ is comparing the first pair of class labels
# with 9 classes, there are 9 * 8 / 2 distinct
coef_img = masker.inverse_transform(model_svm.coef_[0, :])
plotting.view_img(
coef_img, bg_img=haxby_dataset.anat[0],
title="SVM weights", dim=-1, resampling_interpolation='nearest'
)

