An overview of the Haxby dataset#
This part of the tutorial aims to make participants familiar with the dataset we are going to use during this session and also address/introduce/recap some important aspects concerning datasets within machine learning/decoding. The objectives 📍 are:
(re-)familiarize everyone with important
datasetsaspectsexploring and understand the
tutorial dataset
A short primer on datasets#
We wanted to avoid “just talking” about brain decoding in theory and also showcase how the respective models and workflows can be implemented, as well as run to give you some first hands-on experience. Even though we would have loved to get everyone to bring their data and directly apply the things we talk about, it’s unfortunately a bit too time-consuming for this setting. Thus, we decided to utilize an example dataset that is ready to go and “small enough” to run decoding models locally, ie on laptops. You might think “One of those tutorials again…it works with the example dataset but I have little or no chance on running it on/adapting it to my data.” and we would agree based on workshops we did ourselves.
However, we tried our best to address this here by utilizing software whose workflows and processing steps are rather agnostic and implemented via high-level API that should allow a comparably straightforward application to different kinds of data. This specifically refers to a set of core aspects concerning the dataset’s structure and information entailed therein. How about a brief recap?
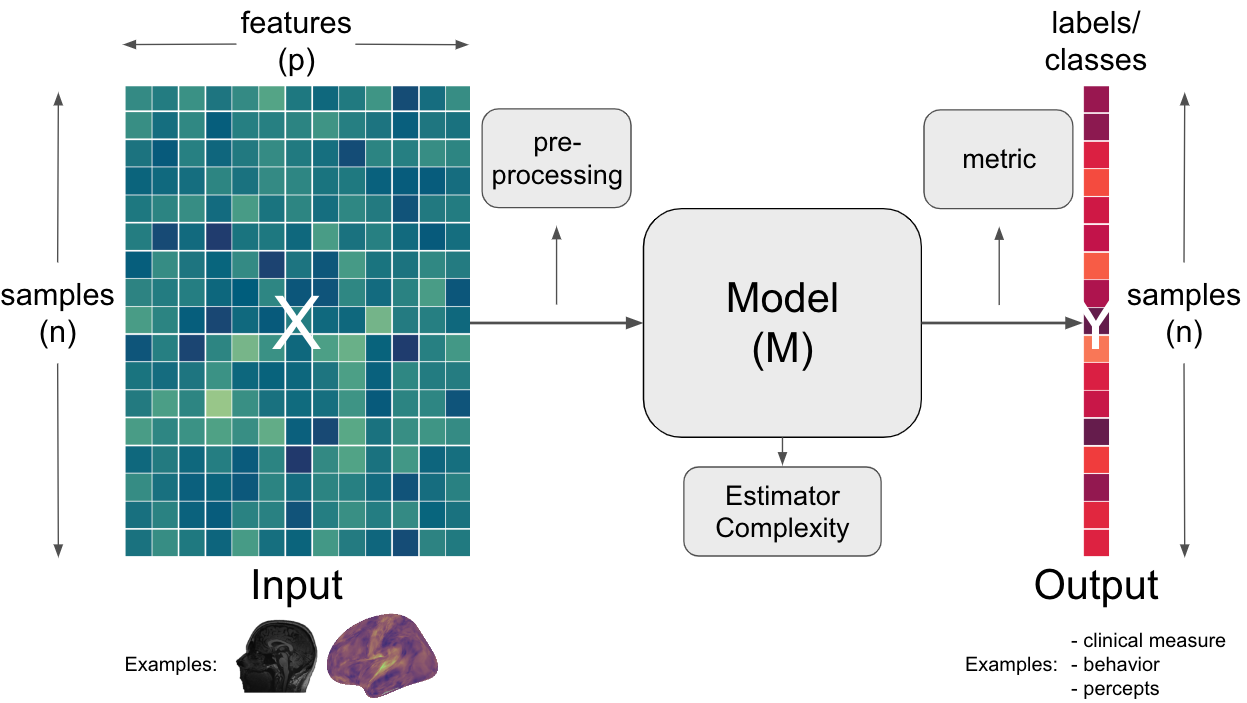
Fig. 3 A schematic representation of standard decoding workflow/pipeline. The input (data) is prepared and potentially preprocessed before being submitted to a model that then utilizes a certain metric to provide a certain output.#
Here, we are going to focus on the input (data). As you heard before, it is usually expected to be structured as samples X features.
What could samples X features refer to/entail?
A sample could be considered an observation/data point/one distinct entity in the dataset/one distinct part of the dataset. For example, if you want to predict what a participant perceived based on their brain activation/response, the samples could entail the fMRI scans or estimated contrast images of that participant. If you want to predict whether a participant exhibited a certain behavior, e.g. a captured by a clinical measure, etc., then the samples could comprise different participants.
A feature on the other hand would entail/describe certain aspects of a given sample. For example, if you want to predict what a participant perceived based on their brain activation/response, the features could entail the voxel pattern at a certain ROI at the given sample.
Thus, in order to make a given dataset “ready” for machine learning/decoding, we need to get it into the respective structure. Lucky for us, the tools we are going to explore, specifically nilearn, incorporate this aspect and are make the corresponding process rather easy. What you need to run machine learning/decoding on your dataset is:
know what your
samplesare (e.g.time series,statistical maps, etc.)know what your
featuresare (e.g.voxel patternof anROI,annotations, etc.)get the
datasetin the formsamplesXfeatures, iesamplesarerowsandfeaturesarecolumns
While exploring the tutorial dataset we will refer to this to make it more clear.
Bonus question: ever heard of the “small-n-high-p” (p >> n) problem?
“Classical” machine learning/decoding models and the underlying algorithms operate on the assumption that are more samples than there are predictors or features . In fact many more. Why is that?
Consider a high-dimensional space whose dimensions are defined by the number of features (e.g. 10 features would result in a space with 10 dimensions. The resulting volume of this space is the amount of samples that could be drawn from the domain and the number of samples entail the samples you need to address your learning problem, ie decoding outcome. That is why folks say: “get more data”, machine learning is data-hungry: our sample needs to be as representative of the high-dimensional domain as possible. Thus, as the number of features increases, so should the number of samples so to capture enough of the space for the decoding model at hand.
This referred to as the curse of dimensionality and poses as a major problem in many fields that aim to utilize machine learning/decoding on unsuitable data. Why is that?
Just imagine we have way more features than samples, ie 50 features and 10 samples. Instead of having a large amount of samples within the space, allowing to achieve a sufficient coverage of the latter, we now have a very high-dimensional space (50 dimensions) and only very few samples therein, basically not allowing us to capture nearly enough of the space as we would need to. This can result in expected outcomes, misleading results or even lead to complete model failure. Furthermore, respective datasets often lead to models that are overfitted and don’t generalize well.
However, there are a few things that can be done to address this, including feature selection, projection into lower-dimensional spaces or representations or regularization.
Question for everyone: what kind have datasets do we usually have in neuroscience, especially neuroimaging?
Downloading & exploring the Haxby dataset#
In the field of functional magnetic resonance imaging (fMRI), one of the first studies which have demonstrated the feasibility of brain decoding was the study by Haxby and colleagues (2001) [HGF+01]. Subjects were presented with various images drawn from different categories and subsequently a decoding model used to predict the presented categories based on the brain activity/responses. In the respective parts of this session, we will try to do the same!
We are going to start with one subject, number 4. To get the data, we can simply use nilearn’s dataset module. At first, we need to import the respective module.
import os
from nilearn import datasets
Next, we get the data and going to save it in a directory called data. Depending on your machine and internet connection, this might take a minute or so.
data_dir = os.path.join('..', 'data')
haxby_dataset = datasets.fetch_haxby(subjects=[4], fetch_stimuli=True, data_dir=data_dir)
What do we have now? Lets have a look!
haxby_dataset
{'anat': ['../data/haxby2001/subj4/anat.nii.gz'],
'func': ['../data/haxby2001/subj4/bold.nii.gz'],
'session_target': ['../data/haxby2001/subj4/labels.txt'],
'mask_vt': ['../data/haxby2001/subj4/mask4_vt.nii.gz'],
'mask_face': ['../data/haxby2001/subj4/mask8b_face_vt.nii.gz'],
'mask_house': ['../data/haxby2001/subj4/mask8b_house_vt.nii.gz'],
'mask_face_little': ['../data/haxby2001/subj4/mask8_face_vt.nii.gz'],
'mask_house_little': ['../data/haxby2001/subj4/mask8_house_vt.nii.gz'],
'mask': '../data/haxby2001/mask.nii.gz',
'description': '.. _haxby_dataset:\n\nHaxby dataset\n=============\n\nAccess\n------\nSee :func:`nilearn.datasets.fetch_haxby`.\n\nNotes\n-----\nResults from a classical :term:`fMRI` study that investigated the differences between\nthe neural correlates of face versus object processing in the ventral visual\nstream. Face and object stimuli showed widely distributed and overlapping\nresponse patterns.\n\nSee :footcite:t:`Haxby2001`.\n\nContent\n-------\nThe "simple" dataset includes:\n :\'func\': Nifti images with bold data\n :\'session_target\': Text file containing run data\n :\'mask\': Nifti images with employed mask\n :\'session\': Text file with condition labels\n\nThe full dataset additionally includes\n :\'anat\': Nifti images with anatomical image\n :\'func\': Nifti images with bold data\n :\'mask_vt\': Nifti images with mask for ventral visual/temporal cortex\n :\'mask_face\': Nifti images with face-reponsive brain regions\n :\'mask_house\': Nifti images with house-reponsive brain regions\n :\'mask_face_little\': Spatially more constrained version of the above\n :\'mask_house_little\': Spatially more constrained version of the above\n\nReferences\n----------\n\n.. footbibliography::\n\nFor more information see:\nPyMVPA provides a tutorial using this dataset :\nhttp://www.pymvpa.org/tutorial.html\n\nMore information about its structure :\nhttp://dev.pymvpa.org/datadb/haxby2001.html\n\n\nLicense\n-------\nunknown\n',
'stimuli': {'shoes': ['../data/haxby2001/stimuli/shoes/shoea1.jpg',
'../data/haxby2001/stimuli/shoes/shoea2.jpg',
'../data/haxby2001/stimuli/shoes/shoea3.jpg',
'../data/haxby2001/stimuli/shoes/shoea5.jpg',
'../data/haxby2001/stimuli/shoes/shoeb1.jpg',
'../data/haxby2001/stimuli/shoes/shoeb2.jpg',
'../data/haxby2001/stimuli/shoes/shoeb4.jpg',
'../data/haxby2001/stimuli/shoes/shoec1.jpg',
'../data/haxby2001/stimuli/shoes/shoec2.jpg',
'../data/haxby2001/stimuli/shoes/shoec3.jpg',
'../data/haxby2001/stimuli/shoes/shoec5.jpg',
'../data/haxby2001/stimuli/shoes/shoed1.jpg',
'../data/haxby2001/stimuli/shoes/shoed2.jpg',
'../data/haxby2001/stimuli/shoes/shoed3.jpg',
'../data/haxby2001/stimuli/shoes/shoed5.jpg',
'../data/haxby2001/stimuli/shoes/shoee1.jpg',
'../data/haxby2001/stimuli/shoes/shoee2.jpg',
'../data/haxby2001/stimuli/shoes/shoee3.jpg',
'../data/haxby2001/stimuli/shoes/shoee5.jpg',
'../data/haxby2001/stimuli/shoes/shoef1.jpg',
'../data/haxby2001/stimuli/shoes/shoef2.jpg',
'../data/haxby2001/stimuli/shoes/shoef3.jpg',
'../data/haxby2001/stimuli/shoes/shoef5.jpg',
'../data/haxby2001/stimuli/shoes/shoeg1.jpg',
'../data/haxby2001/stimuli/shoes/shoeg2.jpg',
'../data/haxby2001/stimuli/shoes/shoeg3.jpg',
'../data/haxby2001/stimuli/shoes/shoeg4.jpg',
'../data/haxby2001/stimuli/shoes/shoeh1.jpg',
'../data/haxby2001/stimuli/shoes/shoeh2.jpg',
'../data/haxby2001/stimuli/shoes/shoeh3.jpg',
'../data/haxby2001/stimuli/shoes/shoeh4.jpg',
'../data/haxby2001/stimuli/shoes/shoei1.jpg',
'../data/haxby2001/stimuli/shoes/shoei2.jpg',
'../data/haxby2001/stimuli/shoes/shoei3.jpg',
'../data/haxby2001/stimuli/shoes/shoei4.jpg',
'../data/haxby2001/stimuli/shoes/shoep1.jpg',
'../data/haxby2001/stimuli/shoes/shoep2.jpg',
'../data/haxby2001/stimuli/shoes/shoep3.jpg',
'../data/haxby2001/stimuli/shoes/shoep4.jpg',
'../data/haxby2001/stimuli/shoes/shoeu1.jpg',
'../data/haxby2001/stimuli/shoes/shoeu2.jpg',
'../data/haxby2001/stimuli/shoes/shoeu3.jpg',
'../data/haxby2001/stimuli/shoes/shoeu4.jpg',
'../data/haxby2001/stimuli/shoes/shoev1.jpg',
'../data/haxby2001/stimuli/shoes/shoev2.jpg',
'../data/haxby2001/stimuli/shoes/shoev3.jpg',
'../data/haxby2001/stimuli/shoes/shoev4.jpg'],
'cats': ['../data/haxby2001/stimuli/cats/MISTY3.jpg',
'../data/haxby2001/stimuli/cats/MISTY4.jpg',
'../data/haxby2001/stimuli/cats/MISTY5.jpg',
'../data/haxby2001/stimuli/cats/MISTY6.jpg',
'../data/haxby2001/stimuli/cats/SPOTZ1.jpg',
'../data/haxby2001/stimuli/cats/SPOTZ4.jpg',
'../data/haxby2001/stimuli/cats/SPOTZ5.jpg',
'../data/haxby2001/stimuli/cats/SPOTZ8.jpg',
'../data/haxby2001/stimuli/cats/brenda1.jpg',
'../data/haxby2001/stimuli/cats/brenda2.jpg',
'../data/haxby2001/stimuli/cats/brenda4.jpg',
'../data/haxby2001/stimuli/cats/brenda5.jpg',
'../data/haxby2001/stimuli/cats/bugs4.jpg',
'../data/haxby2001/stimuli/cats/bugs5.jpg',
'../data/haxby2001/stimuli/cats/bugs7.jpg',
'../data/haxby2001/stimuli/cats/bugs8.jpg',
'../data/haxby2001/stimuli/cats/lucky12.jpg',
'../data/haxby2001/stimuli/cats/lucky13.jpg',
'../data/haxby2001/stimuli/cats/lucky4.jpg',
'../data/haxby2001/stimuli/cats/lucky7.jpg',
'../data/haxby2001/stimuli/cats/majellan1.jpg',
'../data/haxby2001/stimuli/cats/majellan2.jpg',
'../data/haxby2001/stimuli/cats/majellan3.jpg',
'../data/haxby2001/stimuli/cats/majellan4.jpg',
'../data/haxby2001/stimuli/cats/mickey1.jpg',
'../data/haxby2001/stimuli/cats/mickey2.jpg',
'../data/haxby2001/stimuli/cats/mickey3.jpg',
'../data/haxby2001/stimuli/cats/mickey4.jpg',
'../data/haxby2001/stimuli/cats/orange1.jpg',
'../data/haxby2001/stimuli/cats/orange2.jpg',
'../data/haxby2001/stimuli/cats/orange3.jpg',
'../data/haxby2001/stimuli/cats/orange4.jpg',
'../data/haxby2001/stimuli/cats/pepper1.jpg',
'../data/haxby2001/stimuli/cats/pepper2.jpg',
'../data/haxby2001/stimuli/cats/pepper3.jpg',
'../data/haxby2001/stimuli/cats/pepper5.jpg',
'../data/haxby2001/stimuli/cats/robo1.jpg',
'../data/haxby2001/stimuli/cats/robo2.jpg',
'../data/haxby2001/stimuli/cats/robo4.jpg',
'../data/haxby2001/stimuli/cats/robo5.jpg',
'../data/haxby2001/stimuli/cats/stripes2.jpg',
'../data/haxby2001/stimuli/cats/stripes3.jpg',
'../data/haxby2001/stimuli/cats/stripes5.jpg',
'../data/haxby2001/stimuli/cats/stripes6.jpg',
'../data/haxby2001/stimuli/cats/wookie6.jpg',
'../data/haxby2001/stimuli/cats/wookie7.jpg',
'../data/haxby2001/stimuli/cats/wookie8.jpg',
'../data/haxby2001/stimuli/cats/wookie9.jpg'],
'houses': ['../data/haxby2001/stimuli/houses/house1.1.jpg',
'../data/haxby2001/stimuli/houses/house1.2.jpg',
'../data/haxby2001/stimuli/houses/house1.3.jpg',
'../data/haxby2001/stimuli/houses/house1.4.jpg',
'../data/haxby2001/stimuli/houses/house10.1.jpg',
'../data/haxby2001/stimuli/houses/house10.2.jpg',
'../data/haxby2001/stimuli/houses/house10.3.jpg',
'../data/haxby2001/stimuli/houses/house10.4.jpg',
'../data/haxby2001/stimuli/houses/house11.1.jpg',
'../data/haxby2001/stimuli/houses/house11.2.jpg',
'../data/haxby2001/stimuli/houses/house11.3.jpg',
'../data/haxby2001/stimuli/houses/house11.4.jpg',
'../data/haxby2001/stimuli/houses/house12.1.jpg',
'../data/haxby2001/stimuli/houses/house12.2.jpg',
'../data/haxby2001/stimuli/houses/house12.3.jpg',
'../data/haxby2001/stimuli/houses/house12.4.jpg',
'../data/haxby2001/stimuli/houses/house2.1.jpg',
'../data/haxby2001/stimuli/houses/house2.2.jpg',
'../data/haxby2001/stimuli/houses/house2.3.jpg',
'../data/haxby2001/stimuli/houses/house2.4.jpg',
'../data/haxby2001/stimuli/houses/house3.1.jpg',
'../data/haxby2001/stimuli/houses/house3.2.jpg',
'../data/haxby2001/stimuli/houses/house3.3.jpg',
'../data/haxby2001/stimuli/houses/house3.4.jpg',
'../data/haxby2001/stimuli/houses/house4.1.jpg',
'../data/haxby2001/stimuli/houses/house4.2.jpg',
'../data/haxby2001/stimuli/houses/house4.3.jpg',
'../data/haxby2001/stimuli/houses/house4.4.jpg',
'../data/haxby2001/stimuli/houses/house5.1.jpg',
'../data/haxby2001/stimuli/houses/house5.2.jpg',
'../data/haxby2001/stimuli/houses/house5.3.jpg',
'../data/haxby2001/stimuli/houses/house5.4.jpg',
'../data/haxby2001/stimuli/houses/house6.1.jpg',
'../data/haxby2001/stimuli/houses/house6.2.jpg',
'../data/haxby2001/stimuli/houses/house6.3.jpg',
'../data/haxby2001/stimuli/houses/house6.4.jpg',
'../data/haxby2001/stimuli/houses/house7.1.jpg',
'../data/haxby2001/stimuli/houses/house7.2.jpg',
'../data/haxby2001/stimuli/houses/house7.3.jpg',
'../data/haxby2001/stimuli/houses/house7.4.jpg',
'../data/haxby2001/stimuli/houses/house8.1.jpg',
'../data/haxby2001/stimuli/houses/house8.2.jpg',
'../data/haxby2001/stimuli/houses/house8.3.jpg',
'../data/haxby2001/stimuli/houses/house8.4.jpg',
'../data/haxby2001/stimuli/houses/house9.1.jpg',
'../data/haxby2001/stimuli/houses/house9.2.jpg',
'../data/haxby2001/stimuli/houses/house9.3.jpg',
'../data/haxby2001/stimuli/houses/house9.4.jpg'],
'chairs': ['../data/haxby2001/stimuli/chairs/d23a.jpg',
'../data/haxby2001/stimuli/chairs/d23b.jpg',
'../data/haxby2001/stimuli/chairs/d23c.jpg',
'../data/haxby2001/stimuli/chairs/d23d.jpg',
'../data/haxby2001/stimuli/chairs/d25a.jpg',
'../data/haxby2001/stimuli/chairs/d25b.jpg',
'../data/haxby2001/stimuli/chairs/d25c.jpg',
'../data/haxby2001/stimuli/chairs/d25d.jpg',
'../data/haxby2001/stimuli/chairs/d30a.jpg',
'../data/haxby2001/stimuli/chairs/d30b.jpg',
'../data/haxby2001/stimuli/chairs/d30c.jpg',
'../data/haxby2001/stimuli/chairs/d30d.jpg',
'../data/haxby2001/stimuli/chairs/d37a.jpg',
'../data/haxby2001/stimuli/chairs/d37b.jpg',
'../data/haxby2001/stimuli/chairs/d37c.jpg',
'../data/haxby2001/stimuli/chairs/d37d.jpg',
'../data/haxby2001/stimuli/chairs/d38a.jpg',
'../data/haxby2001/stimuli/chairs/d38b.jpg',
'../data/haxby2001/stimuli/chairs/d38c.jpg',
'../data/haxby2001/stimuli/chairs/d38d.jpg',
'../data/haxby2001/stimuli/chairs/d39a.jpg',
'../data/haxby2001/stimuli/chairs/d39b.jpg',
'../data/haxby2001/stimuli/chairs/d39c.jpg',
'../data/haxby2001/stimuli/chairs/d39d.jpg',
'../data/haxby2001/stimuli/chairs/d62a.jpg',
'../data/haxby2001/stimuli/chairs/d62b.jpg',
'../data/haxby2001/stimuli/chairs/d62c.jpg',
'../data/haxby2001/stimuli/chairs/d62d.jpg',
'../data/haxby2001/stimuli/chairs/d63a.jpg',
'../data/haxby2001/stimuli/chairs/d63b.jpg',
'../data/haxby2001/stimuli/chairs/d63c.jpg',
'../data/haxby2001/stimuli/chairs/d63d.jpg',
'../data/haxby2001/stimuli/chairs/d67a.jpg',
'../data/haxby2001/stimuli/chairs/d67b.jpg',
'../data/haxby2001/stimuli/chairs/d67c.jpg',
'../data/haxby2001/stimuli/chairs/d67d.jpg',
'../data/haxby2001/stimuli/chairs/d79a.jpg',
'../data/haxby2001/stimuli/chairs/d79b.jpg',
'../data/haxby2001/stimuli/chairs/d79c.jpg',
'../data/haxby2001/stimuli/chairs/d79d.jpg',
'../data/haxby2001/stimuli/chairs/d85a.jpg',
'../data/haxby2001/stimuli/chairs/d85b.jpg',
'../data/haxby2001/stimuli/chairs/d85c.jpg',
'../data/haxby2001/stimuli/chairs/d85d.jpg',
'../data/haxby2001/stimuli/chairs/d9a.jpg',
'../data/haxby2001/stimuli/chairs/d9b.jpg',
'../data/haxby2001/stimuli/chairs/d9c.jpg',
'../data/haxby2001/stimuli/chairs/d9d.jpg'],
'scissors': ['../data/haxby2001/stimuli/scissors/scissor1.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor1.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor1.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor1.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor10.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor10.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor10.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor10.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor11.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor11.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor11.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor11.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor12.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor12.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor12.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor12.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor2.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor2.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor2.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor2.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor3.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor3.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor3.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor3.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor4.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor4.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor4.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor4.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor5.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor5.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor5.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor5.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor6.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor6.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor6.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor6.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor7.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor7.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor7.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor7.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor8.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor8.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor8.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor8.4.jpg',
'../data/haxby2001/stimuli/scissors/scissor9.1.jpg',
'../data/haxby2001/stimuli/scissors/scissor9.2.jpg',
'../data/haxby2001/stimuli/scissors/scissor9.3.jpg',
'../data/haxby2001/stimuli/scissors/scissor9.4.jpg'],
'faces': ['../data/haxby2001/stimuli/faces/Annie_1.jpg',
'../data/haxby2001/stimuli/faces/Annie_2.jpg',
'../data/haxby2001/stimuli/faces/Annie_3.jpg',
'../data/haxby2001/stimuli/faces/Annie_4.jpg',
'../data/haxby2001/stimuli/faces/Blake_1.jpg',
'../data/haxby2001/stimuli/faces/Blake_2.jpg',
'../data/haxby2001/stimuli/faces/Blake_3.jpg',
'../data/haxby2001/stimuli/faces/Blake_4.jpg',
'../data/haxby2001/stimuli/faces/Don_1.jpg',
'../data/haxby2001/stimuli/faces/Don_2.jpg',
'../data/haxby2001/stimuli/faces/Don_3.jpg',
'../data/haxby2001/stimuli/faces/Don_4.jpg',
'../data/haxby2001/stimuli/faces/Estelle_1.jpg',
'../data/haxby2001/stimuli/faces/Estelle_2.jpg',
'../data/haxby2001/stimuli/faces/Estelle_3.jpg',
'../data/haxby2001/stimuli/faces/Estelle_4.jpg',
'../data/haxby2001/stimuli/faces/Frank_1.jpg',
'../data/haxby2001/stimuli/faces/Frank_2.jpg',
'../data/haxby2001/stimuli/faces/Frank_3.jpg',
'../data/haxby2001/stimuli/faces/Frank_4.jpg',
'../data/haxby2001/stimuli/faces/Janie_1.jpg',
'../data/haxby2001/stimuli/faces/Janie_2.jpg',
'../data/haxby2001/stimuli/faces/Janie_3.jpg',
'../data/haxby2001/stimuli/faces/Janie_4.jpg',
'../data/haxby2001/stimuli/faces/Joan_1.jpg',
'../data/haxby2001/stimuli/faces/Joan_2.jpg',
'../data/haxby2001/stimuli/faces/Joan_3.jpg',
'../data/haxby2001/stimuli/faces/Joan_4.jpg',
'../data/haxby2001/stimuli/faces/Jodi_1.jpg',
'../data/haxby2001/stimuli/faces/Jodi_2.jpg',
'../data/haxby2001/stimuli/faces/Jodi_3.jpg',
'../data/haxby2001/stimuli/faces/Jodi_4.jpg',
'../data/haxby2001/stimuli/faces/Joe_1.jpg',
'../data/haxby2001/stimuli/faces/Joe_2.jpg',
'../data/haxby2001/stimuli/faces/Joe_3.jpg',
'../data/haxby2001/stimuli/faces/Joe_4.jpg',
'../data/haxby2001/stimuli/faces/Tim_1.jpg',
'../data/haxby2001/stimuli/faces/Tim_2.jpg',
'../data/haxby2001/stimuli/faces/Tim_3.jpg',
'../data/haxby2001/stimuli/faces/Tim_4.jpg',
'../data/haxby2001/stimuli/faces/Tom_1.jpg',
'../data/haxby2001/stimuli/faces/Tom_2.jpg',
'../data/haxby2001/stimuli/faces/Tom_3.jpg',
'../data/haxby2001/stimuli/faces/Tom_4.jpg',
'../data/haxby2001/stimuli/faces/Wallace_1.jpg',
'../data/haxby2001/stimuli/faces/Wallace_2.jpg',
'../data/haxby2001/stimuli/faces/Wallace_3.jpg',
'../data/haxby2001/stimuli/faces/Wallace_4.jpg'],
'bottles': ['../data/haxby2001/stimuli/bottles/bottle1.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle1.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle1.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle1.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle10.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle10.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle10.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle10.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle11.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle11.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle11.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle11.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle12.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle12.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle12.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle12.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle2.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle2.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle2.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle2.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle3.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle3.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle3.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle3.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle4.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle4.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle4.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle4.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle5.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle5.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle5.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle5.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle6.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle6.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle6.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle6.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle7.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle7.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle7.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle7.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle8.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle8.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle8.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle8.4.jpg',
'../data/haxby2001/stimuli/bottles/bottle9.1.jpg',
'../data/haxby2001/stimuli/bottles/bottle9.2.jpg',
'../data/haxby2001/stimuli/bottles/bottle9.3.jpg',
'../data/haxby2001/stimuli/bottles/bottle9.4.jpg'],
'controls': [('scrambled_bottles',
['../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle1.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle1.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle1.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle1.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle10.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle10.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle10.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle10.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle11.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle11.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle11.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle11.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle12.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle12.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle12.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle12.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle2.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle2.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle2.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle2.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle3.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle3.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle3.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle3.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle4.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle4.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle4.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle4.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle5.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle5.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle5.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle5.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle6.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle6.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle6.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle6.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle7.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle7.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle7.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle7.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle8.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle8.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle8.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle8.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle9.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle9.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle9.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_bottles/scrambled_bottle9.4.jpg']),
('scrambled_cats',
['../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_MISTY3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_MISTY4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_MISTY5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_MISTY6.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_SPOTZ1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_SPOTZ4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_SPOTZ5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_SPOTZ8.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_brenda1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_brenda2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_brenda4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_brenda5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_bugs4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_bugs5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_bugs7.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_bugs8.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_lucky12.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_lucky13.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_lucky4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_lucky7.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_majellan1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_majellan2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_majellan3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_majellan4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_mickey1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_mickey2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_mickey3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_mickey4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_orange1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_orange2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_orange3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_orange4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_pepper1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_pepper2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_pepper3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_pepper5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_robo1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_robo2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_robo4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_robo5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_stripes2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_stripes3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_stripes5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_stripes6.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_wookie6.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_wookie7.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_wookie8.jpg',
'../data/haxby2001/stimuli/controls/scrambled_cats/scrambled_wookie9.jpg']),
('scrambled_chairs',
['../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d23a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d23b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d23c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d23d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d25a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d25b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d25c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d25d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d30a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d30b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d30c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d30d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d37a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d37b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d37c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d37d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d38a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d38b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d38c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d38d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d39a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d39b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d39c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d39d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d62a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d62b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d62c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d62d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d63a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d63b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d63c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d63d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d67a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d67b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d67c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d67d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d79a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d79b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d79c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d79d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d85a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d85b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d85c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d85d.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d9a.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d9b.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d9c.jpg',
'../data/haxby2001/stimuli/controls/scrambled_chairs/scrambled_d9d.jpg']),
('scrambled_faces',
['../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Annie_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Annie_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Annie_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Annie_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Blake_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Blake_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Blake_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Blake_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Don_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Don_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Don_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Don_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Estelle_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Estelle_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Estelle_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Estelle_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Frank_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Frank_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Frank_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Frank_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Janie_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Janie_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Janie_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Janie_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Joan_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Joan_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Joan_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Joan_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Jodi_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Jodi_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Jodi_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Jodi_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Joe_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Joe_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Joe_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Joe_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Tim_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Tim_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Tim_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Tim_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Tom_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Tom_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Tom_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Tom_4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Wallace_1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Wallace_2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Wallace_3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_faces/scrambled_Wallace_4.jpg']),
('scrambled_houses',
['../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house1.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house1.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house1.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house1.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house10.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house10.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house10.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house10.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house2.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house2.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house2.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house2.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house3.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house3.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house3.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house3.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house4.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house4.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house4.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house4.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house5.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house5.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house5.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house5.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house6.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house6.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house6.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house6.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house7.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house7.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house7.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house7.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house8.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house8.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house8.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house8.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house9.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house9.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house9.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_houses/scrambled_house9.4.jpg']),
('scrambled_scissors',
['../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor1.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor1.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor1.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor1.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor10.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor10.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor10.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor10.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor11.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor11.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor11.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor11.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor12.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor12.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor12.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor12.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor2.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor2.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor2.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor2.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor3.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor3.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor3.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor3.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor4.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor4.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor4.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor4.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor5.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor5.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor5.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor5.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor6.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor6.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor6.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor6.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor7.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor7.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor7.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor7.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor8.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor8.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor8.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor8.4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor9.1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor9.2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor9.3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_scissors/scrambled_scissor9.4.jpg']),
('scrambled_shoes',
['../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoea1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoea2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoea3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoea5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeb1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeb2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeb3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeb4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoec1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoec2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoec3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoec5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoed1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoed2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoed3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoed5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoee1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoee2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoee3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoee5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoef1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoef2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoef3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoef5.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeg1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeg2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeg3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeg4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeh1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeh2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeh3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeh4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoei1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoei2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoei3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoei4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoep1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoep2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoep3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoep4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeu1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeu2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeu3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoeu4.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoev1.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoev2.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoev3.jpg',
'../data/haxby2001/stimuli/controls/scrambled_shoes/scrambled_shoev4.jpg'])]}}
As you can see, we get a python dictionary and there’s quite bit in it. This includes:
the
anatomical dataunderanatthe
functional dataunderfuncan annotation when
participantsperceived whatcategoryseveral
masksundermask*a
datasetdescriptionstimuli categoriesand respectivestimuli
Thinking about input data again…
What would be our samples and features?
The data in more depth#
After getting a first idea of what our dataset entails, we should spend a bit more time exploring it in more depth, starting with the neuroimaging files.
Neuroimaging files#
As seen above, the data includes several nii files, which contain images of brain volumes, either anatomical or functional scans, as well as (binary) masks. Lets have a look at the anatomical image first.
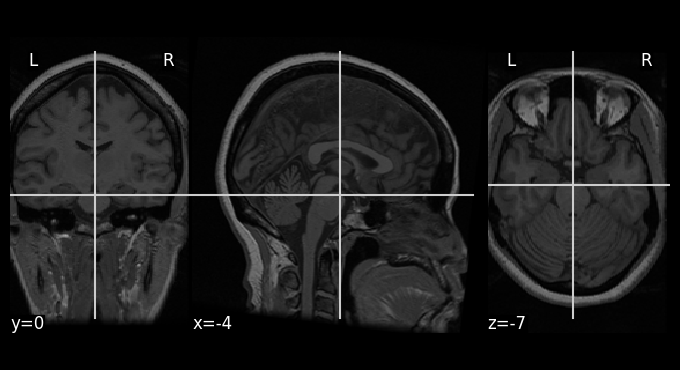
Using nilearn, we can either load and then plot it or directly plot it. Here we are going to do the first option as it will allow us to check the properties of the image.
from nilearn.image import load_img
anat_image = load_img(haxby_dataset.anat)
Now we can access basically all parts of the image, including the header
print(anat_image.header)
<class 'nibabel.nifti1.Nifti1Header'> object, endian='<'
sizeof_hdr : 348
data_type : b''
db_name : b''
extents : 0
session_error : 0
regular : b'r'
dim_info : 0
dim : [ 4 124 256 256 1 1 1 1]
intent_p1 : 0.0
intent_p2 : 0.0
intent_p3 : 0.0
intent_code : none
datatype : int16
bitpix : 16
slice_start : 0
pixdim : [1. 1.2 0.9375 0.9375 1. 1. 1. 1. ]
vox_offset : 0.0
scl_slope : nan
scl_inter : nan
slice_end : 0
slice_code : unknown
xyzt_units : 10
cal_max : 1007.0
cal_min : 0.0
slice_duration : 0.0
toffset : 0.0
glmax : 0
glmin : 0
descrip : b'FSL3.3'
aux_file : b''
qform_code : unknown
sform_code : unknown
quatern_b : 0.0
quatern_c : 0.0
quatern_d : 0.0
qoffset_x : 0.0
qoffset_y : 0.0
qoffset_z : 0.0
srow_x : [0. 0. 0. 0.]
srow_y : [0. 0. 0. 0.]
srow_z : [0. 0. 0. 0.]
intent_name : b''
magic : b'n+1'
and actual data.
anat_image.dataobj
array([[[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
...,
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]]],
[[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
...,
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]]],
[[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
...,
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]]],
...,
[[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
...,
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]]],
[[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
...,
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]]],
[[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
...,
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]],
[[0],
[0],
[0],
...,
[0],
[0],
[0]]]], dtype=int16)
anat_image.dataobj.shape
(124, 256, 256, 1)
As you can see, this is basically a numpy array that has the same dimensions as our image and the data reflect values for a given voxel. So far so good but how does it actually look? We can make use of one of nilearn’s many plotting functions.
from nilearn import plotting
plotting.plot_anat(anat_image)
<nilearn.plotting.displays._slicers.OrthoSlicer at 0x7f0b9cbd7650>
We can even create an interactive plot:
plotting.view_img(anat_image, symmetric_cmap=False, cmap='Greys_r', colorbar=False)
/opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/numpy/core/fromnumeric.py:771: UserWarning: Warning: 'partition' will ignore the 'mask' of the MaskedArray.
a.partition(kth, axis=axis, kind=kind, order=order)
/opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/nilearn/image/resampling.py:756: UserWarning: Casting data from int32 to float32
return resample_img(
Comparably, we can do the same things with the functional image. That is loading the image:
func_image = load_img(haxby_dataset.func)
and inspect its header:
print(func_image.header)
<class 'nibabel.nifti1.Nifti1Header'> object, endian='<'
sizeof_hdr : 348
data_type : b''
db_name : b''
extents : 0
session_error : 0
regular : b'r'
dim_info : 0
dim : [ 4 40 64 64 1452 1 1 1]
intent_p1 : 0.0
intent_p2 : 0.0
intent_p3 : 0.0
intent_code : none
datatype : int16
bitpix : 16
slice_start : 0
pixdim : [1. 3.5 3.75 3.75 2.5 0. 0. 0. ]
vox_offset : 0.0
scl_slope : nan
scl_inter : nan
slice_end : 0
slice_code : unknown
xyzt_units : 10
cal_max : 3312.0
cal_min : 0.0
slice_duration : 0.0
toffset : 0.0
glmax : 0
glmin : 0
descrip : b'FSL4.0'
aux_file : b''
qform_code : unknown
sform_code : unknown
quatern_b : 0.0
quatern_c : 0.0
quatern_d : 0.0
qoffset_x : 0.0
qoffset_y : 0.0
qoffset_z : 0.0
srow_x : [0. 0. 0. 0.]
srow_y : [0. 0. 0. 0.]
srow_z : [0. 0. 0. 0.]
intent_name : b''
magic : b'n+1'
and data:
func_image.get_fdata()
array([[[[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[10., 11., 10., ..., 0., 0., 0.],
[16., 28., 14., ..., 0., 0., 0.],
[17., 23., 28., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 13., 7., 8.],
[ 0., 0., 0., ..., 21., 18., 13.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[ 4., 11., 11., ..., 0., 0., 0.],
[17., 26., 16., ..., 0., 0., 0.],
[35., 32., 35., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 24., 14., 11.],
[ 0., 0., 0., ..., 22., 24., 23.],
[ 0., 0., 0., ..., 0., 0., 0.]],
...,
[[ 0., 0., 0., ..., 0., 0., 0.],
[10., 11., 17., ..., 0., 0., 0.],
[21., 14., 29., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 10., 14., 10.],
[ 0., 0., 0., ..., 4., 7., 4.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 0., 0., 0.],
[ 8., 13., 15., ..., 0., 0., 0.],
[23., 21., 31., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 9., 7., 11.],
[ 0., 0., 0., ..., 6., 3., 3.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 0., 0., 0.],
[10., 28., 21., ..., 0., 0., 0.],
[17., 35., 36., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 12., 15., 13.],
[ 0., 0., 0., ..., 7., 13., 9.],
[ 0., 0., 0., ..., 0., 0., 0.]]],
[[[ 0., 0., 0., ..., 18., 24., 13.],
[ 0., 0., 0., ..., 20., 24., 24.],
[ 0., 0., 0., ..., 34., 33., 25.],
...,
[15., 24., 18., ..., 0., 0., 0.],
[ 7., 9., 11., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[10., 9., 14., ..., 7., 24., 25.],
[14., 14., 14., ..., 17., 28., 26.],
[12., 14., 21., ..., 15., 17., 35.],
...,
[ 8., 16., 18., ..., 19., 8., 9.],
[ 9., 7., 10., ..., 12., 9., 17.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[ 8., 6., 10., ..., 20., 23., 25.],
[12., 6., 14., ..., 22., 24., 21.],
[22., 18., 16., ..., 21., 18., 43.],
...,
[20., 28., 17., ..., 11., 26., 16.],
[ 9., 12., 8., ..., 14., 14., 8.],
[ 0., 0., 0., ..., 0., 0., 0.]],
...,
[[ 0., 0., 0., ..., 23., 18., 12.],
[20., 23., 32., ..., 28., 37., 14.],
[30., 22., 32., ..., 24., 37., 11.],
...,
[22., 17., 23., ..., 22., 10., 17.],
[22., 19., 15., ..., 5., 4., 6.],
[ 7., 9., 12., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 12., 24., 17.],
[17., 14., 17., ..., 32., 43., 25.],
[20., 9., 17., ..., 36., 37., 20.],
...,
[19., 34., 20., ..., 13., 15., 10.],
[20., 33., 21., ..., 6., 10., 6.],
[17., 22., 18., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 0., 0., 0.],
[18., 22., 19., ..., 0., 0., 0.],
[29., 39., 24., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 15., 16., 20.],
[ 0., 0., 0., ..., 6., 7., 9.],
[ 0., 0., 0., ..., 0., 0., 0.]]],
[[[ 0., 0., 0., ..., 21., 29., 28.],
[ 0., 0., 0., ..., 23., 34., 29.],
[ 0., 0., 0., ..., 22., 49., 29.],
...,
[18., 15., 21., ..., 0., 0., 0.],
[12., 3., 10., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[21., 23., 16., ..., 13., 8., 11.],
[28., 23., 11., ..., 17., 20., 19.],
[29., 35., 19., ..., 7., 21., 55.],
...,
[25., 23., 14., ..., 12., 17., 35.],
[15., 6., 10., ..., 22., 18., 13.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[12., 16., 17., ..., 16., 14., 11.],
[20., 15., 16., ..., 11., 7., 14.],
[32., 44., 11., ..., 5., 18., 40.],
...,
[31., 34., 23., ..., 2., 16., 18.],
[18., 12., 20., ..., 9., 10., 19.],
[ 0., 0., 0., ..., 0., 0., 0.]],
...,
[[ 0., 0., 0., ..., 24., 26., 44.],
[20., 21., 23., ..., 21., 28., 45.],
[21., 24., 20., ..., 22., 31., 28.],
...,
[14., 27., 21., ..., 19., 17., 32.],
[20., 29., 22., ..., 4., 11., 16.],
[11., 17., 12., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 23., 25., 38.],
[21., 20., 21., ..., 24., 24., 46.],
[20., 13., 20., ..., 23., 16., 31.],
...,
[29., 25., 24., ..., 25., 32., 47.],
[20., 30., 18., ..., 12., 18., 21.],
[15., 21., 15., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 0., 0., 0.],
[14., 13., 20., ..., 0., 0., 0.],
[19., 12., 23., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 31., 32., 46.],
[ 0., 0., 0., ..., 13., 19., 19.],
[ 0., 0., 0., ..., 0., 0., 0.]]],
...,
[[[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 32., 13., 20.],
[ 0., 0., 0., ..., 15., 22., 41.],
...,
[26., 37., 20., ..., 0., 0., 0.],
[16., 18., 17., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[23., 26., 34., ..., 0., 0., 0.],
[37., 28., 31., ..., 43., 30., 41.],
[37., 28., 34., ..., 12., 20., 27.],
...,
[26., 21., 20., ..., 22., 17., 20.],
[22., 11., 24., ..., 25., 19., 10.],
[ 0., 0., 0., ..., 1., 0., 0.]],
[[12., 10., 28., ..., 0., 24., 0.],
[31., 18., 23., ..., 31., 32., 29.],
[25., 23., 19., ..., 20., 30., 33.],
...,
[34., 26., 22., ..., 30., 15., 35.],
[32., 17., 18., ..., 18., 18., 20.],
[ 0., 0., 0., ..., 0., 0., 0.]],
...,
[[ 0., 0., 0., ..., 20., 12., 7.],
[31., 24., 34., ..., 33., 16., 19.],
[36., 26., 37., ..., 22., 24., 26.],
...,
[18., 28., 19., ..., 19., 21., 21.],
[22., 23., 19., ..., 8., 9., 9.],
[ 9., 13., 15., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 27., 22., 10.],
[34., 20., 29., ..., 34., 16., 22.],
[35., 20., 32., ..., 23., 20., 38.],
...,
[32., 40., 18., ..., 18., 21., 17.],
[29., 32., 12., ..., 5., 12., 12.],
[12., 20., 13., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 0., 0., 0.],
[31., 26., 29., ..., 0., 0., 0.],
[32., 30., 24., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 29., 19., 21.],
[ 0., 0., 0., ..., 14., 4., 11.],
[ 0., 0., 0., ..., 0., 0., 0.]]],
[[[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 32., 19., 29.],
[ 0., 0., 0., ..., 27., 16., 32.],
...,
[20., 27., 21., ..., 0., 0., 0.],
[14., 20., 17., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[23., 24., 18., ..., 0., 0., 0.],
[39., 24., 20., ..., 25., 26., 32.],
[29., 40., 32., ..., 33., 17., 28.],
...,
[17., 18., 20., ..., 31., 9., 38.],
[10., 10., 13., ..., 16., 18., 11.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[16., 16., 13., ..., 0., 10., 0.],
[24., 26., 19., ..., 29., 18., 29.],
[27., 32., 20., ..., 24., 20., 25.],
...,
[21., 12., 16., ..., 13., 11., 25.],
[14., 9., 16., ..., 24., 11., 8.],
[ 0., 0., 0., ..., 0., 0., 0.]],
...,
[[ 0., 0., 0., ..., 18., 16., 14.],
[11., 16., 13., ..., 22., 16., 16.],
[19., 23., 11., ..., 13., 14., 17.],
...,
[24., 28., 14., ..., 24., 11., 6.],
[20., 18., 10., ..., 16., 11., 2.],
[10., 8., 7., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 17., 24., 28.],
[16., 13., 16., ..., 12., 30., 26.],
[27., 22., 18., ..., 16., 31., 26.],
...,
[21., 31., 16., ..., 18., 20., 17.],
[22., 31., 15., ..., 11., 17., 12.],
[11., 21., 15., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 0., 0., 0.],
[18., 24., 19., ..., 0., 0., 0.],
[26., 16., 18., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 17., 18., 16.],
[ 0., 0., 0., ..., 13., 17., 9.],
[ 0., 0., 0., ..., 0., 0., 0.]]],
[[[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 26., 14., 19.],
[ 0., 0., 0., ..., 30., 21., 21.],
...,
[22., 24., 12., ..., 0., 0., 0.],
[ 7., 14., 6., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 35., 9., 20.],
[ 0., 0., 0., ..., 24., 18., 20.],
...,
[25., 20., 18., ..., 0., 0., 0.],
[10., 10., 7., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 0., 7., 0.],
[ 0., 0., 0., ..., 20., 24., 11.],
[ 0., 0., 0., ..., 16., 30., 21.],
...,
[28., 17., 17., ..., 0., 0., 0.],
[16., 10., 12., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.]],
...,
[[ 0., 0., 0., ..., 14., 20., 16.],
[ 0., 0., 0., ..., 19., 29., 15.],
[ 0., 0., 0., ..., 18., 29., 21.],
...,
[22., 19., 24., ..., 0., 0., 0.],
[11., 13., 13., ..., 0., 0., 0.],
[ 6., 6., 9., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 13., 12., 13.],
[ 0., 0., 0., ..., 17., 12., 16.],
[ 0., 0., 0., ..., 15., 15., 25.],
...,
[24., 24., 23., ..., 0., 0., 0.],
[ 9., 16., 17., ..., 0., 0., 0.],
[ 5., 5., 9., ..., 0., 0., 0.]],
[[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.],
...,
[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.]]]])
func_image.dataobj.shape
(40, 64, 64, 1452)
We already noticed something…
The data of the anatomical and functional image are quite different. Do you know why and which we would use for our planned decoding analyses?
As we have a 4D image, that is brain volumes acquired over time (the 4th dimension), we need to adapt the plotting a bit. More precisely, we need to either plot a 3D image at a given time point or e.g. compute the mean image over time and plot that. The latter might be more informative and additional shows you how easy this can be done using nilearn’s image functions. Thus, we, at first, import the respective function and compute the mean image:
from nilearn.image import mean_img
func_image_mean = mean_img(func_image)
We can check if this worked via the approach we followed above, ie checking the data:
func_image_mean.dataobj.shape
(40, 64, 64)
That seems about right and we can give the plot a try:
plotting.plot_epi(func_image_mean, cmap='magma')
<nilearn.plotting.displays._slicers.OrthoSlicer at 0x7f0b9cb78250>
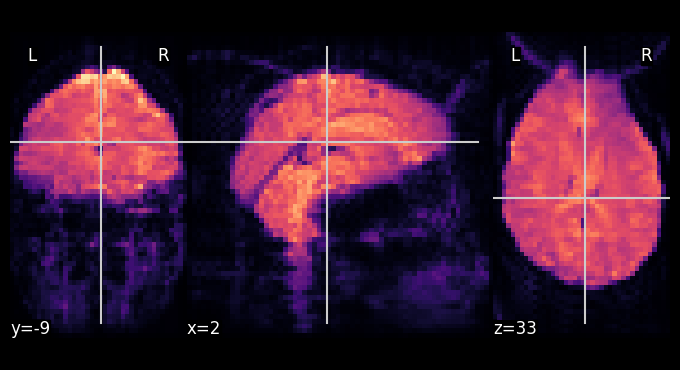
and of course, this also works for interactive plots.
plotting.view_img(func_image_mean, cmap='magma', symmetric_cmap=False)
/opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/numpy/core/fromnumeric.py:771: UserWarning: Warning: 'partition' will ignore the 'mask' of the MaskedArray.
a.partition(kth, axis=axis, kind=kind, order=order)
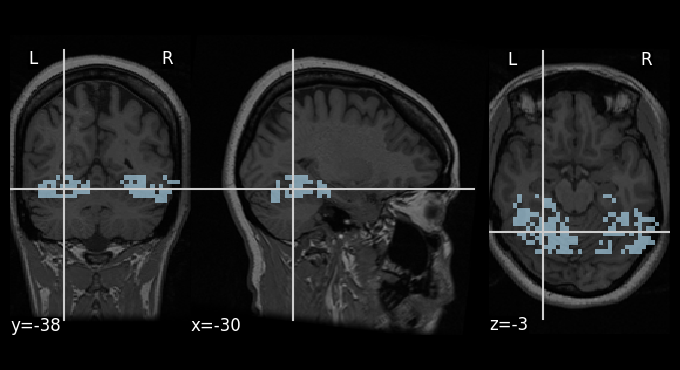
The last type of neuroimaging file we need to check are the (binary) masks, so let’s do it for one example mask: the ventral temporal cortex. This mask has been generated as part of the Haxby et al. (2001) study [HGF+01], and highlights a part of the brain specialized in the processing of visual information, and which contains areas sensitive to different types of image categories [GSW14] . As with the types before, we can load,
vt_mask = load_img(haxby_dataset.mask_vt)
inspect
print(vt_mask.header)
<class 'nibabel.nifti1.Nifti1Header'> object, endian='<'
sizeof_hdr : 348
data_type : b''
db_name : b''
extents : 0
session_error : 0
regular : b'r'
dim_info : 0
dim : [ 4 40 64 64 1 1 1 1]
intent_p1 : 0.0
intent_p2 : 0.0
intent_p3 : 0.0
intent_code : none
datatype : float32
bitpix : 32
slice_start : 0
pixdim : [1. 3.5 3.75 3.75 1. 1. 1. 1. ]
vox_offset : 0.0
scl_slope : nan
scl_inter : nan
slice_end : 0
slice_code : unknown
xyzt_units : 10
cal_max : 1.0
cal_min : 0.0
slice_duration : 0.0
toffset : 0.0
glmax : 0
glmin : 0
descrip : b'FSL3.3'
aux_file : b''
qform_code : unknown
sform_code : unknown
quatern_b : 0.0
quatern_c : 0.0
quatern_d : 0.0
qoffset_x : 0.0
qoffset_y : 0.0
qoffset_z : 0.0
srow_x : [0. 0. 0. 0.]
srow_y : [0. 0. 0. 0.]
srow_z : [0. 0. 0. 0.]
intent_name : b''
magic : b'n+1'
vt_mask.get_fdata()
array([[[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
...,
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]]],
[[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
...,
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]]],
[[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
...,
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]]],
...,
[[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
...,
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]]],
[[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
...,
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]]],
[[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
...,
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]],
[[0.],
[0.],
[0.],
...,
[0.],
[0.],
[0.]]]])
vt_mask.dataobj.shape
(40, 64, 64, 1)
and visualize it (Here, we are going to plot it as an overlay on the anatomical image).
plotting.plot_roi(vt_mask, bg_img=anat_image,
cmap='Paired')
<nilearn.plotting.displays._slicers.OrthoSlicer at 0x7f0b9aa31210>
With that, we had a quick look at all neuroimaging file types present in the dataset and can continue to have a look at the other file types (and information therein) required to apply a decoding model.
Labels and stimulus annotations#
As mentioned in prior sessions (e.g.Supervised learning using scikit-learn and hinted at the beginning of this session), when working on a supervised learning problem, we also need the ground truth/true labels for each sample. Why? Because we need to evaluate how a given model performs via comparing the labels it predicted to the true labels. What these labels refer to can be manifold and of course depends on the task at hand.
For example, a supervised learning problem in the dataset at hand could entail training a model to recognize and predict what category participants perceived based on their brain activation. Thus, we would need to know what category was shown when during the acquisition of the data (or which category resulted in which estimated brain activity).
Within our tutorial dataset, this information is included in the session_target file. Using pandas we can easily load and inspect this file:
import pandas as pd
stimulus_annotations = pd.read_csv(haxby_dataset.session_target[0], delimiter=' ')
stimulus_annotations.head(n=200)
| labels | chunks | |
|---|---|---|
| 0 | rest | 0 |
| 1 | rest | 0 |
| 2 | rest | 0 |
| 3 | rest | 0 |
| 4 | rest | 0 |
| ... | ... | ... |
| 195 | rest | 1 |
| 196 | rest | 1 |
| 197 | rest | 1 |
| 198 | rest | 1 |
| 199 | shoe | 1 |
200 rows × 2 columns
While this is already informative, let’s plot it to get a better intuition.
import seaborn as sns
import matplotlib.pyplot as plt
ax = sns.scatterplot(x=stimulus_annotations.index, y=stimulus_annotations['labels'],
hue=stimulus_annotations['labels'], legend=False, palette='colorblind')
plt.title('Categories shown across time')
ax.set_xlabel('Time point/fMRI scan')
sns.despine(offset=5)
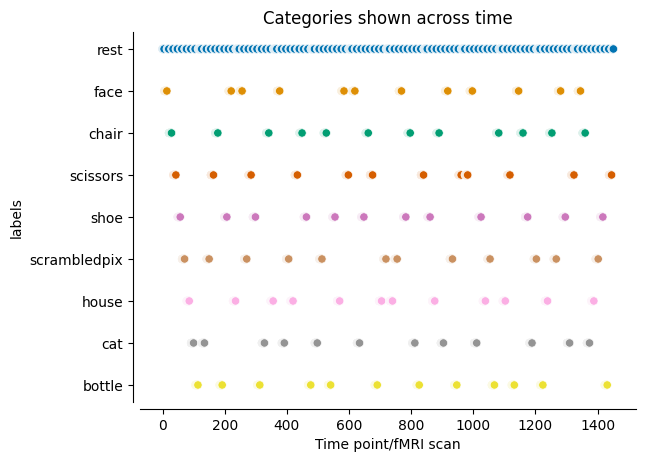
As we can see, the information provided indicates what category participants perceived at which sample or fMRI image acquisition aka point in time during the experiment. With that, we have the needed labels for our samples (ie our Y) and can thus apply a supervised learning problem.
Summary#
This already concludes this section of the session within which we explored the went through basic datasets concepts again and afterwards explored the tutorial dataset which we are going to use during the remaining sections of this session, ie Decoding via SVM, Decoding using MLPs and Decoding using GCNs.
Within this section you should have learned:
important aspects of
datasetsstructured input in the form
samples X featuressmall n high pproblem
the
tutorial datasetbackground
file types and information therein
neuroimagingfilesstimulus annotations
If you have any questions, please don’t hesitate to ask us. Thank you very much for your attention and see you in the next section.
References#
Kalanit Grill-Spector and Kevin S. Weiner. The functional architecture of the ventral temporal cortex and its role in categorization. Nature Reviews Neuroscience, 15(8):536–548, August 2014. URL: https://doi.org/10.1038/nrn3747, doi:10.1038/nrn3747.
Bonus: checking the stimuli#
As you saw above, our tutorial dataset actually also contains the stimuli utilized in the experiment. This pretty unique (because of e.g. copyright problems) but really cool. As we could use the stimuli for certain analyses, e.g. encoding and/or comparing their processing in biological and artificial neural networks. However, this is unfortunately outside the scope of this session. Thus, we are just going to plot a few of them so you get an impression.
We can examine one functional volume using nilearn’s plotting tools. Because fmri data are 4D we use nilearn.image.mean_img to extract the average brain volume.
import matplotlib.pyplot as plt
from nilearn import datasets
from nilearn.plotting import show
stimulus_information = haxby_dataset.stimuli
for stim_type in stimulus_information:
# skip control images, there are too many
if stim_type != 'controls':
file_names = stimulus_information[stim_type]
file_names = file_names[0:16]
fig, axes = plt.subplots(4, 4)
fig.suptitle(stim_type)
for img_path, ax in zip(file_names, axes.ravel()):
ax.imshow(plt.imread(img_path), cmap=plt.cm.gray)
for ax in axes.ravel():
ax.axis("off")
show()



Please note that for each image category, a number of scrambled images were also presented.
for stim_num in range(len(stimulus_information['controls'])):
stim_type = stimulus_information['controls'][stim_num][0]
file_names = stimulus_information['controls'][stim_num][1]
file_names = file_names[0:16]
fig, axes = plt.subplots(4, 4)
fig.suptitle(stim_type)
for img_path, ax in zip(file_names, axes.ravel()):
ax.imshow(plt.imread(img_path), cmap=plt.cm.gray)
for ax in axes.ravel():
ax.axis("off")
show()