Welcome#
“Introduction to brain decoding in fMRI”
This jupyter book presents an introduction to brain decoding using fMRI. It was developed within the educational courses, conducted as part of the Montreal AI and Neuroscience (MAIN) conference in October 2024.
Building upon the prior sections of the educational courses, the here presented resources aim to provide an overview of how decoding models can be applied to fMRI data in order to investigate brain function. Importantly, the respective methods cannot only be utilized to analyze data from biological agents (e.g. humans, non-human primates, etc.) but also artificial neural networks, as well as presenting the opportunity to compare processing in both. They are thus core approaches that are prominently used at the intersection of neuroscience and AI.
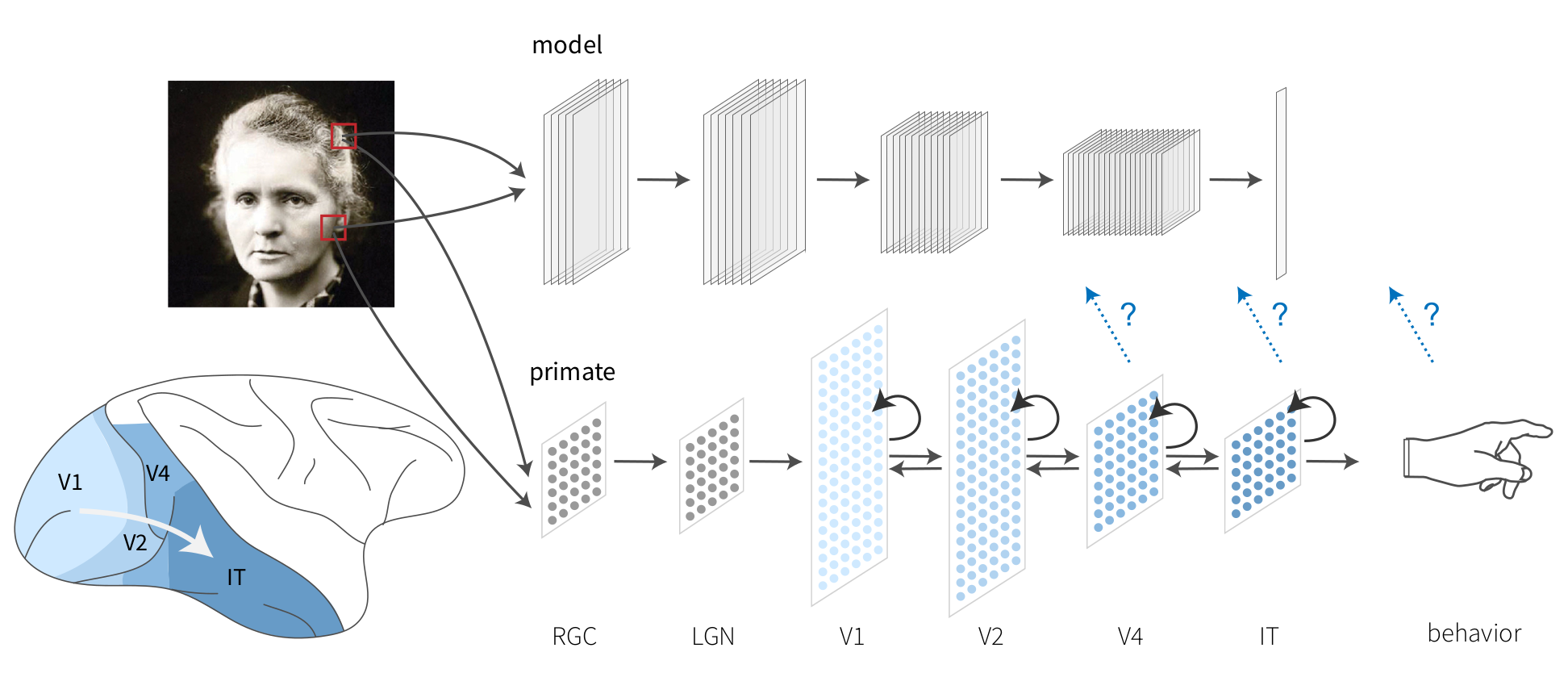
Fig. 1 To test the consistency of representations in artificial neural networks (ANNs) and the brain, it is possible to encode brain activity based on ANN presented with similar stimuli, or decode brain activity by predicting the expected ANN activity and corresponding annotation of cognitive states. Figure from Schrimpf et al. (2020) [SKH+20], under a CC-BY 4.0 license.#
The tutorials make heavy use of nilearn concerning
manipulating and processing fMRI data, as well as scikit-learn and pytorch to apply decoding models on the data.
We used the Jupyter Book framework to provide all materials in an open, structured and interactive manner. ALl pages and section you see here are built from markdown files or jupyter notebooks, allowing you to read through the materials and/or run them, locally or in the cloud. The three symbols on the top right allow to enable full screen mode, link to the underlying GitHub repository and allow you to download the respective sections as a pdf or jupyter notebook respectively. Some sections will additionally have a little rocket in that row which will allow you to interactively rerun certain parts via cloud computing (please see the Binder section for more information).
Brain decoding vs. encoding#
In short, encoding and decoding entail contrary operations that can yet be utilized in a complementary manner. Encoding models applied to brain data, e.g. fMRI, aim to predict brain responses/activity based on annotations or features of the stimuli perceived by the participant. These can be obtained from a multitude of options, including artificial neural networks which would allow to relate their processing of the stimuli to that of biological agents, ie brains.
Decoding models on the other hand comprise models with which we aim to estimate/predict what a participant is perceiving or doing based on recordings of brain responses/activity, e.g. fMRI.
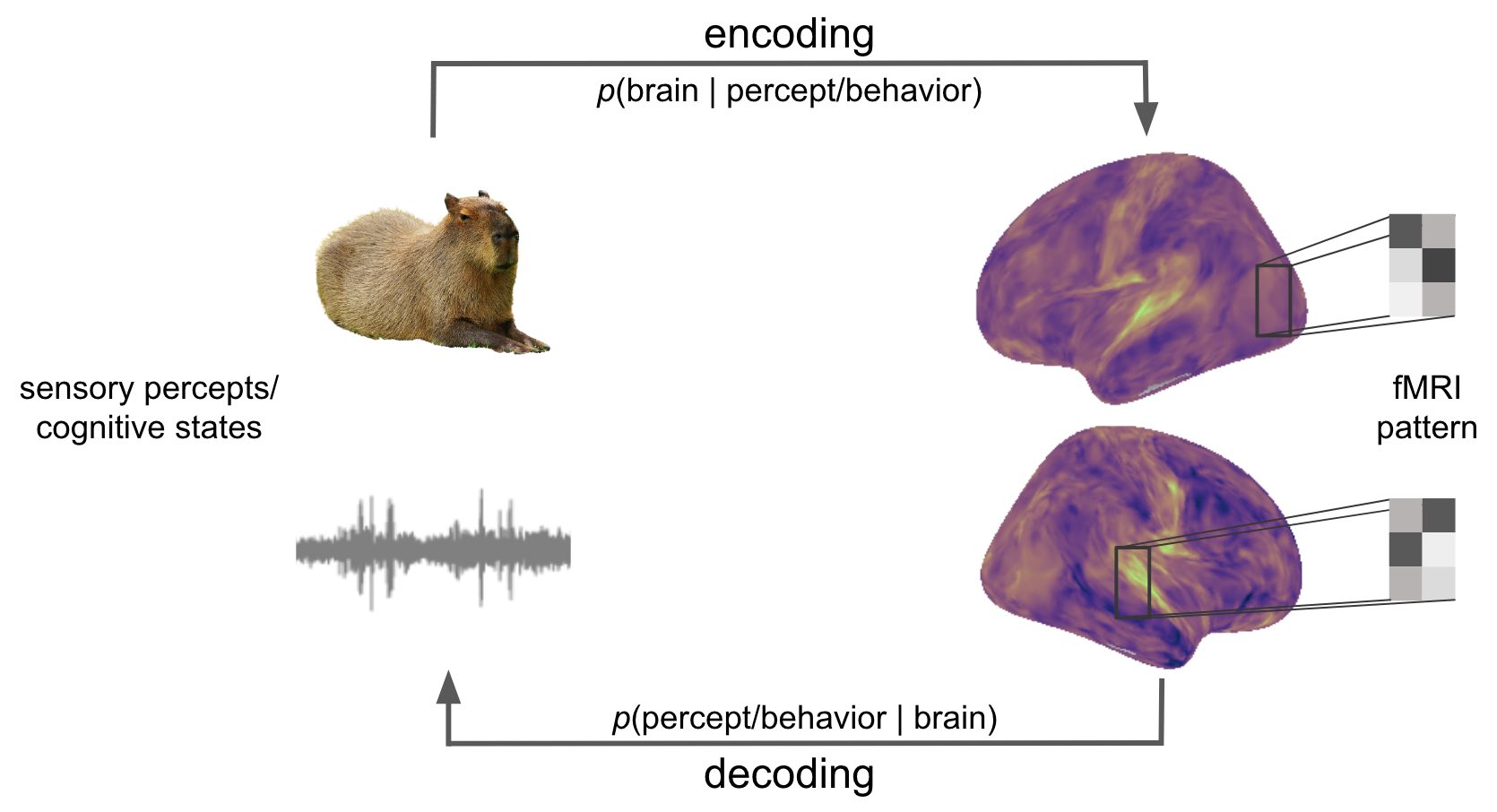
Fig. 2 Encoding and decoding present contrary, yet complementary operations. While the former targets the prediction of brain activity/responses based on stimulus percepts/features (e.g. vision & audition), cognitive states or behavior, the latter aims to predict those aspects based on brain activity/responses.#
More information and their application can be found in the respective sections of this resource. You can either use the ToC on the left or the links below to navigate accordingly.
Setup#
There are two ways to run the tutorials: local installation and using free cloud computing provided by Binder. As noted below, we strongly recommend the local installation, as the Binder option comes with limited computational resources, as well as the missed possibility to directly further explore the approaches presented in this tutorial on your own machine.
For the local installation to work, you need two things: the fitting python environment and the content. Concerning python, please have a look at the hint below.
Install python
You need to have access to a terminal with Python 3.
If you have setup your environment based on instructions of MAIN educational installation guide, you are good to go 🎉
If it not already the case, here is a quick guide to install python 3 on any OS.
After making sure you have a working python installation, you need to get the content that is going to presented during the tutorial. In more detail, this is done via interactive jupyter notebooks which you can obtain by following the steps below:
Clone/download this repository to your machine and navigate to the directory.
git clone https://github.com/main-educational/brain_encoding_decoding.git cd brain_encoding_decoding
We encourage you to use a
virtual environmentfor this tutorial (and for all your projects, that’s a good practice). To do this, run the following commands in your terminal, it will create theenvironmentin a folder namedmain_edu_brain_decoding:python3 -m venv main_edu_brain_decoding
Then the following
commandwillactivatetheenvironment:source main_edu_brain_decoding/bin/activate
Finally, you can install the required
libraries:pip install -r requirements.txt
Navigate to the
contentof thejupyter book:cd content/
Now that you are all set, you can run the notebooks with the command:
jupyter notebookClick on the
.mdfiles. They will be rendered as jupyter notebooks 🎉
Alternatively, you can use conda/miniconda to create the needed python environment like so:
git clone https://github.com/main-educational/brain_encoding_decoding.git
cd brain_encoding_decoding
conda env create -f environment.yml
If you wish to run the tutorial in Binder, click on the rocket icon 🚀 in the top right of a given notebook to launch it on Binder.
Warning
The computing resource on Binder is limited.
Some cells might not execute correctly, or the data download will not be completed.
For the full experience, we recommend using the local set up instruction.
Instructors#
This tutorial was prepared and presented by
It is based on earlier versions created by:

Thanks and acknowledgements#
Parts of the tutorial are directly adapted from a nilearn tutorial on the so-called Haxby dataset.
It was adapted from a prior version which was prepared and presented by Pravish Sainath Shima Rastegarnia, Hao-Ting Wang Loic Tetrel and Pierre Bellec.
Furthermore, some images and code are used from a previous iteration of this tutorial, prepared by Dr Yu Zhang.
We would like to thank the Jupyter community, specifically, the Executable/Jupyter Book and mybinder project for enabling us to create this tutorial. Furthermore, we are grateful for the entire open neuroscience community and the amazing support and resources it provides. This includes the community driven development of data and processing standards, as well as unbelievable amount of software packages that make the here introduced approaches possible to begin with.
The tutorial is rendered here using Jupyter Book.
References#
Martin Schrimpf, Jonas Kubilius, Ha Hong, Najib J Majaj, Rishi Rajalingham, Elias B Issa, Kohitij Kar, Pouya Bashivan, Jonathan Prescott-Roy, Franziska Geiger, Kailyn Schmidt, Daniel L K Yamins, and James J DiCarlo. Brain-Score: which artificial neural network for object recognition is most Brain-Like? bioRxiv, pages 407007, January 2020.